The Schmidt laboratory: Regulation of cellular lipid homeostasis
Sphingolipids are an essential class of lipids. They function as structural components of all cellular membranes, and are particularly indispensable for myelination in the nervous system. The biosynthesis of sphingolipids is initiated at the endoplasmic reticulum (ER) with the condensation of the amino acid L-serine and palmitic acid by the enzyme serine-palmitoyl-coenzym A transferase (SPT), which forms the first common precursor of all sphingolipids. This reaction is rate-limiting for sphingolipid synthesis and tightly regulated, because accumulation of sphingolipids or especially their synthesis intermediates is detrimental for cells, and associated with a number of diseases frequently including neurodegeneration (e.g. Gaucher’s and Fabri’s disease or hereditary sensory and autonomic neuropathy type 1).
SPT activity is regulated by the evolutionary conserved family of Orm1-like proteins, of which humans possess three paralogues (ORMDL1-3). They are membrane proteins of the ER, which associate with the transmembrane domain of SPT and inhibit its catalytic activity (see Figure A). Why the genomes of most organisms encode more for than one ORMDL paralogue is not understood. Genetic mutations affecting the interaction of ORMDL family proteins with the SPT are of high pathological relevance. Loss of this regulatory module and hence uncontrolled synthesis of sphingolipid precursors is likely causative for a subtype of amyotrophic lateral sclerosis (ALS; Mohassel et al., 2021, Nature medicine), whereas increased expression of ORMDL3 is associated with several common inflammatory diseases, including childhood asthma (Moffatt et al., 2007, Nature) and IBD (MacGovern et al., 2010, Nature genetics).
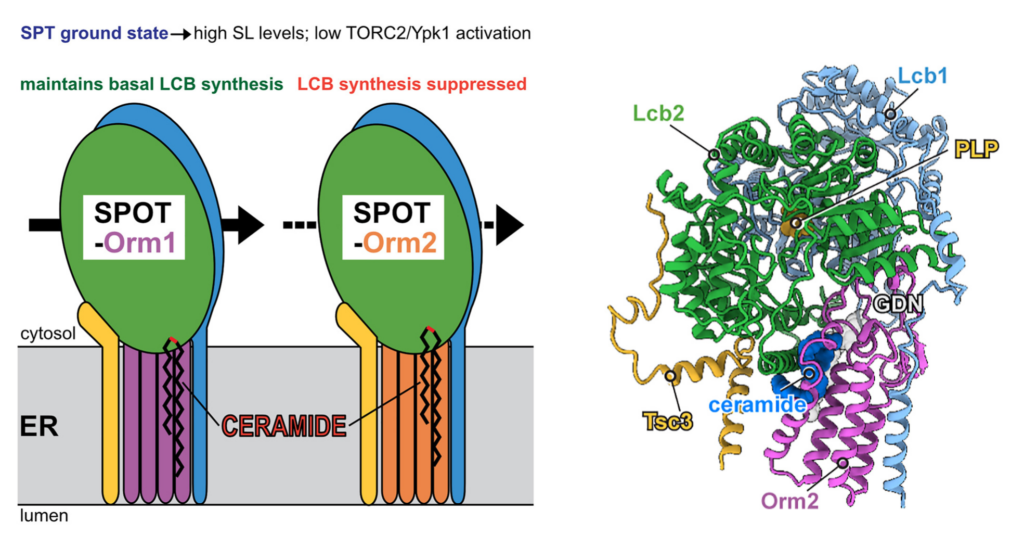
Figure A: Structure of the Orm2-bound, inhibited SPT and different inhibitory potentials of yeast ORMDL proteins Orm1 and Orm2. (adapted from Körner, Schäfer et al., 2024; Cell Reports)
In my lab, we study the regulation of SPT by ORMDL-family proteins particularly in the light of differences between the paralogs, to understand why life typical relies on more than one ORMDL protein. We attempt to address fundamental research questions initially in budding yeast as an easily controllable model system, and subsequently see if they are conserved in humans. We recently found that the inhibitory potential of the two yeast ORMDL proteins, Orm1 and Orm2, is different, with Orm2 intrinsically being the more potent SPT inhibitor (Körner, Schäfer et al., 2024, Cell Rep.).
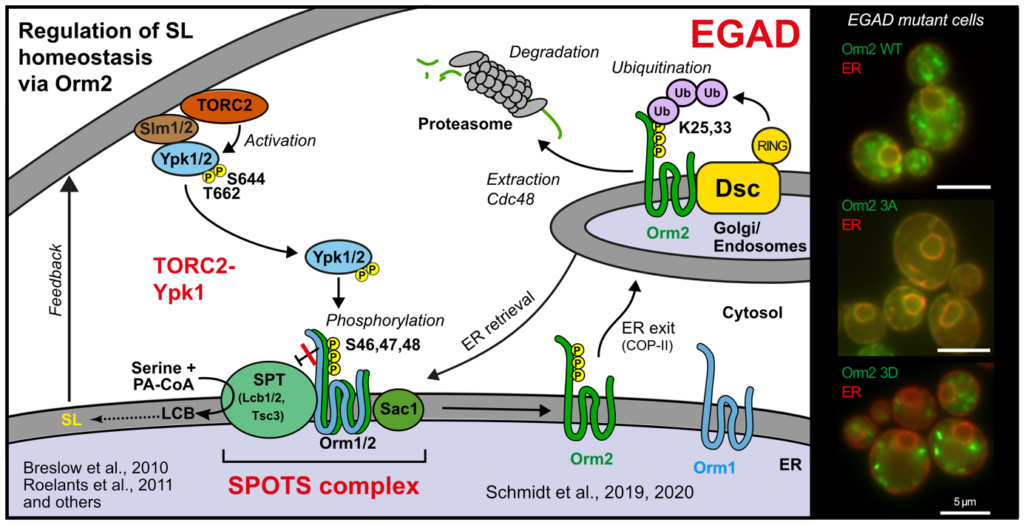
The regulation of SPT by ORMDL proteins is best understood in the budding yeast S. cerevisiae (see Figure B). Orm1 and Orm2 are under the control of plasma membrane stress signaling via the protein kinases TORC2 and Ypk1. Phosphorylation causes dissociation of Orm1/2 and activates the SPT complex (Breslow et al., 2010, Nature), however, this phospho-regulation is only conserved in fungal species. Recently, we described an additional level of regulation of Orm proteins by translocation from the ER and degradation by the endosome and Golgi-associated degradation (EGAD; Schmidt et al., 2019, EMBO J.; Schmidt et al., 2020, JBC). This is the first reported example of a mode of regulation that is not shared by all ORMDL paralogs, because only Orm2 is exported and degraded, while Orm1 is stable. Intriguingly, human ORMDL proteins are also regulated by proteolysis, albeit using a different machinery (Sasset et al., 2023, EMBO Rep.).
Key Publications
The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins. Körner C, Schäfer JH, Esch BM, Parey K, Walter S, Teis D, Januliene D, Schmidt O*, Moeller A*, Fröhlich F*. Cell Rep. 2024 Aug 20;43(8):114627. doi: 10.1016/j.celrep.2024.114627. PMID: 39167489 (* shared corresponding authors)
TOR complex 2 (TORC2) signaling and the ESCRT machinery cooperate in the protection of plasma membrane integrity in yeast. Schmidt O*, Weyer Y, Sprenger S, Widerin MA, Eising S, Baumann V, Angelova M, Loewith R, Stefan CJ, Hess MW, Fröhlich F, Teis D. J Biol Chem. 2020 Aug 21;295(34):12028-12044. doi: 10.1074/jbc.RA120.013222. Epub 2020 Jul 1. PMID: 32611771
Endosome and Golgi-associated degradation (EGAD) of membrane proteins regulates sphingolipid metabolism. Schmidt O, Weyer Y, Baumann V, Widerin MA, Eising S, Angelova M, Schleiffer A, Kremser L, Lindner H, Peter M, Fröhlich F, Teis D. EMBO J. 2019 Aug 1;38(15):e101433. doi: 10.15252/embj.2018101433. Epub 2019 May 27. PMID:31368600
all publications: https://scholar.google.com/citations?user=PHkoQ6cAAAAJ&hl=en
Collaborations
David Teis, MUI Innsbruck, AT
Florian Fröhlich, Osnabrück University, DE
Maria Bohnert, Münster University, DE
Robbie Loewith, University of Geneva, CH
Matthias Peter, ETH Zurich, CH
Funding
Lipotype – Lipid excellence award 2019 (#2)
FWF P36187-B “individual functions of Orm/ORMDL family proteins in membrane homeostasis”
Team and contact

Assoc. Prof. Dr. Oliver Schmidt
Email: oliver.schmidt@i-med.ac.at
Phone: +43 512 9003 70189
Google Scholar: https://scholar.google.com/citations?user=PHkoQ6cAAAAJ&hl=en
ORCID: https://orcid.org/0000-0002-7921-4663
| Name | Position | Contact |
| Oliver Schmidt | group leader | oliver.schmidt@i-med.ac.at |
| Brigitta Seifert | research technician | brigitta.seifert@i-med.ac.at |
| Niklas Schomisch | PhD student | niklas.schomisch@i-med.ac.at |
| Barış Bekdaş | PhD student | baris.bekdas@i-med.ac.at |
| Sophia Pichler | MSc student |
Alumni:
Jana Bleher, MSc student
Bellmunt Blanco, BSc student; University of Barcelona, Erasmus program

June 2024: Lab hike to Sankt Magdalena: from left Niklas, Bariş, Bellmunt, Olli